Abstract
Background
Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL) is an indolent disease. However, a small subset of cases may progress to accelerated CLL (aCLL) and eventually transform to diffuse large B-cell lymphoma, also known as Richter transformation (RT). Core-needle biopsies of CLL in accelerated and transformed phases present a plethora of diagnostic challenges, hindering a confident and precise morphologic assessment. To overcome these impediments, we propose a high-throughput diagnostic pipeline empowered by deep learning to discover and characterize intrinsic cell populations and help boost diagnostic accuracy.
Material & Methods
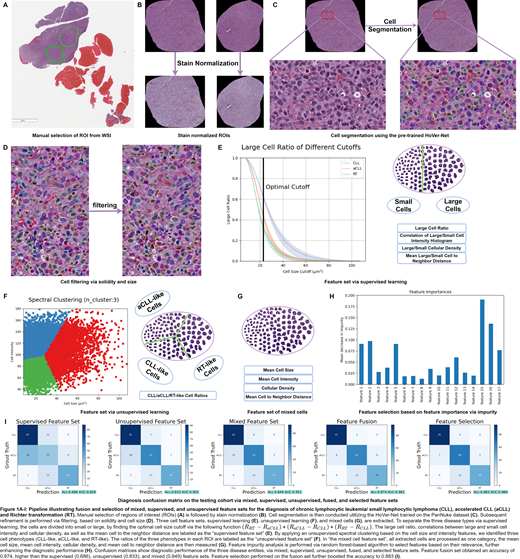
We collected 193 biopsies from 125 patients, including 69 CLL slides from 44 patients, 44 aCLL slides from 34 patients, and 80 RT slides from 47 patients. Our computational pipeline contained the following 7 steps (Figure 1): 1) ROI selection, 2) stain normalization ; 3) cell segmentation through transfer learning, using a pre-trained deep learning model (HoVer-Net); 4) quality control of automated cell segmentation; 5) profiling cell populations from three different perspectives, including grouping of cells into large or small subtypes using supervised learning, discovering the intrinsic cell subpopulations with unsupervised learning, and mixing cells for indiscriminate profiling; 6) pruning uninformative features by quantifying feature importance via impurity analysis; 7) systematically evaluating the diagnostic performance of the three cellular profiling methods as well as feature fusion and selection, followed by two validation strategies: The first one aimed to stratify patients into training and testing cohorts to balance key clinicopathologic factors (one-shot validation); and the second one aimed to randomly split patients into training or testing sets, followed by repeat splitting for 100 times (repeated cross-validation).
Results
First, we sought to define cells into large or small populations, where cell size cutoff was learned to maximize pairwise separation among three disease types (CLL, aCLL, and RT). We then measured large cell ratios, correlations between large and small cell intensity and density, and mean cell to the nearest-neighbor distance, and labeled the extracted attributes as "supervised feature set" (Figure 1E). Second, we applied an unsupervised learning (i.e., spectral clustering) to detect the intrinsic cell subpopulations based on morphology and intensity. Interestingly, three cell phenotypes were uncovered, which we termed as "CLL-like," "aCLL-like," and "RT-like" cells. The ratios of the three cell types in each ROI were computed and labeled as "unsupervised feature set" (Figure 1F). Third, we analyzed cells as one cohort, and computed the mean cell size and intensity, cellular density, and cell to its nearest-neighbor distance, this population was labeled as "mixed cell feature set" as a whole (Figure 1G). lastly, we applied feature selection of fused feature sets, where feature importance was calculated via impurity analysis. Subsequently, 6 out of the total 17 features were pruned (Figure 1H). When testing the three feature sets separately, we observed that the "mixed cell feature set" achieved the best performance (AUC=0.951; n=4 features) followed by the "unsupervised feature set" (AUC=0.902; n=3 features) and "supervised feature set" (AUC=0.829; n=10 features). By integrating the three feature sets, we obtained an accuracy of 0.874 and an area under the curve (AUC) of 0.961 in one-shot validation and a mean accuracy of 0.831 and AUC of 0.952 in repeated cross-validation, surpassing the performance obtained by solely adopting a single feature set at a time. Application of feature selection to fused feature sets further boosted the accuracy to 0.883 and AUC to 0.966 via one-shot validation, and mean accuracy of 0.842 and mean AUC of 0.959 via repeated cross-validation (Figure 1I).
Conclusion
The "mixed feature set" achieved higher diagnostic accuracy in comparison to the "supervised" and "unsupervised" feature sets, emphasizing the power of characterizing heterogeneous cell populations. The synergy of three feature sets validates the hypothesis that integrating different ways of cellular phenotyping may optimize the predictive power. Eliminating less informative features further enhances diagnostic accuracy, highlighting the importance of adopting meaningful attributes.
Khoury: Stemline Therapeutics: Research Funding; Kiromic: Research Funding; Angle: Research Funding.